Editor’s Note: This is the second part of Charles’ introduction to Xgrid. If you are inspired by what you read, don’t forget that we have a prime resource for running your Xgrid jobs right here: OpenMacGrid.
First of all, I want to apologize for the long delay between the first installment and this second installment of the Xgrid tutorial series. In the future, I will try to get better at placing my life priorities on hold to do more of the unpaid fun stuff like writing Xgrid tutorials. Of course, I can not guarantee that GTD won’t get in the way of that ambitious goal.
In the first installment, I introduced you to Xgrid: when to use it and how to use it. One of the main take-home message was that Xgrid was pretty much just like a bunch of well-disciplined graduate students sitting in front of computers and typing commands in the terminal. Except with Xgrid, they are called ‘agents’, you don’t have to pay them, they sit inside the computer, they are invisible, they don’t talk and they don’t eat. They just use the processor when nobody is around.
Table of Contents
The caveman way
In the first installment, we introduced an example of how you could use Xgrid to run sequence alignments on DNA databases, for instance looking for a particular piece of DNA in the human genome. I showed you how you can send jobs to Xgrid, one for each chromosome, and how Xgrid will automatically schedule the jobs and send them to whatever agents are available.
In summary, for each job on a chromosome, you have to:
- submit a job by typing a command like this:
xgrid -job submit /Users/Shared/fasta-tutorial/fasta \ -q /Users/Shared/fasta-tutorial/magic-worm-gene.seq /Users/Shared/fasta-tutorial/chromosomeX.fa - go get a coffee
- check if it is done by typing a command like this:
xgrid -job attributes -id 232 - upload the results
xgrid -job results -id 232 > ~/results-chromosomeX.txt
While relatively straightforward, this process has to be done in parallel for each of the 23 jobs that we need to submit, one for each of the 23 chromosomes (you don’t need to drink 23 coffees, though). We also have to keep track of the job identifiers, which are just meaningless numbers such as -id 232, and remember to which chromosome they correspond. And this is just for the one piece of DNA we are trying to identify in this example, and only running on the 23 chromosomes of the human genome. If you want to run more genes, and scan more genomes, the number of combinations and jobs and IDs will quickly make the process extremely cumbersome and error-prone.
Stuff your grid
An alternative is to use GridStuffer. Disclaimer: I am the developer of GridStuffer, which could make me a bit biased.
My main goal when developing GridStuffer was to provide a tool that would automate much of the repetitive steps listed above. Here are some of the tasks that GridStuffer will perform for you:
- keep track of submissions and make sure every task is completed
- schedule jobs as needed to keep Xgrid fed but not overfed
- retrieve the results automatically and organize them on disk
The best way to see what GridStuffer can do is to put it to work. How about scanning human chromosomes with Fasta?
Get the stuff
First of all, download the latest version of GridStuffer. You may have to first double-click the downloaded zip file. Here is what you will get:
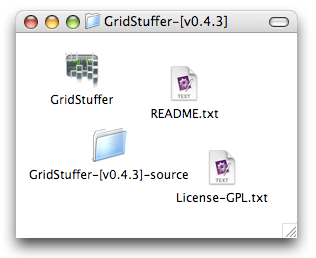
The piece relevant for this tutorial is the GridStuffer application itself, this is the one with the funny-looking icon. I recommand you move it to your Applications folder. Note that there is also a folder with the “source” of GridStuffer. This is the program code that was used to create the application. GridStuffer is open source. If you are an adventurous mac developer, your can see for yourself the guts of GridStuffer. Note however that most of the core Xgrid code is not in GridStuffer, but in a separate framework called GridEZ, also authored by myself, and also open-source.
Connect the stuff
After you double-click the application icon, you should see two windows, named “Controllers” and “Metajobs”. The “Controllers” window is where you will manage connections to Xgrid controllers. In most cases, there should be just one controller listed, and that is your own controller that you started per the instructions in this tutorial. You may actually see more controllers if other people in the building are also playing with Xgrid. Next to the name of the machine, you will see a little orange dot, that indicates that the controller is available, ready to be connected to.
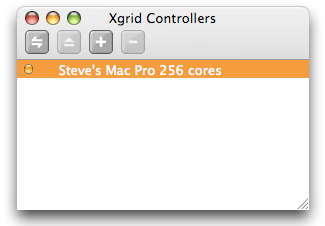
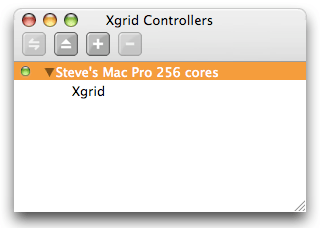
To initiate the connection, select the controller and click on the “Connect” icon (the double arrow on the left in the toolbar), or just press “return” on your keyboard. If you did not set any password, the connection should be almost instantaneous, and the little dot should now be green, indicating that the controller is connected. You should also have a list of the grids managed by this controller. Usually, there is just one grid, interestingly named “Xgrid”. When there are problems with the connection, or a password is required, an additional panel may show up:
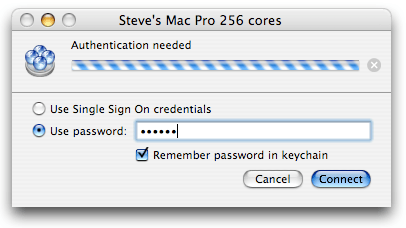
Write the stuff
We now need to prepare a description of your job that GridStuffer can understand. This is in fact very easy, as GridStuffer speaks the same language as the Terminal. All you need to give GridStuffer is the list of commands you want executed on the different machines.
Open TextEdit, and create a new text file by selecting “New” in the “File” menu. Make sure you are creating a plain text file, by choosing “Make Plain Text” in the “Format menu”. In this new file, type the following line:
/Users/Shared/fasta-tutorial/fasta \
-q /Users/Shared/fasta-tutorial/magic-worm-gene.seq /Users/Shared/fasta-tutorial/chromosomeX.fa
This text corresponds exactly to the command you would type in Terminal to run fasta on chromosome X. Now copy and paste this line 22 times. Change the name of the chromosome file to “chromosome1.fa” for the second line, “chromosome2.fa” for the third line, …, “chromosome21.fa” for line 22 and “chromosomeY.fa” for line 23. You should now have a simple list of all the commands that you want to run through Xgrid:
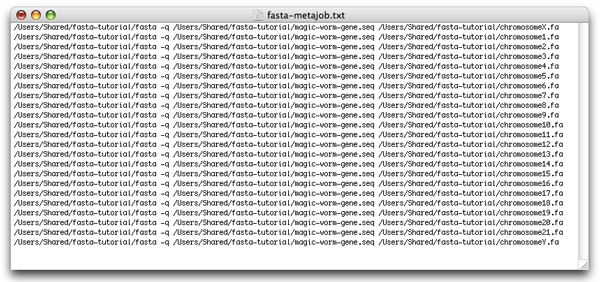
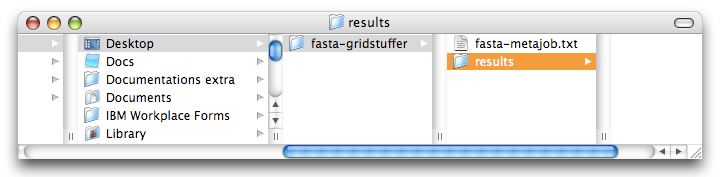
Create a folder on your desktop, called “fasta-gridstuffer”, and save the file you just created under the name “fasta-metajob.txt”. Finally, also, create a folder called “results” inside “fasta-gridstuffer”. You should now see these in the Finder:
Submit the stuff
We are now ready for job submission. Select the “Metajobs” window and click on the ‘+’ icon.
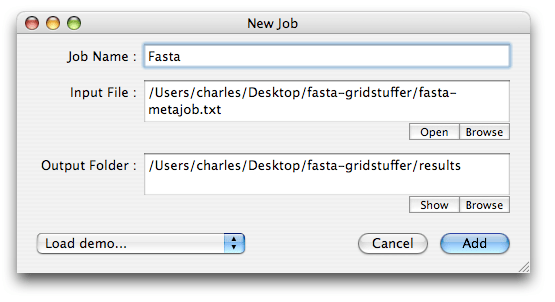
In the “New Job” window that opens, click the ‘Browse’ buttons to show GridStuffer where the input file is (what a coincidence, this is the file we just created), and where you want to store the results (this is the folder we also just created). You may give a name to this metajob, for instance “Fasta”. When all is set, click the ‘Add’ button.
To start the submission process, select the newly created metajob, and click the Start button (arrow icon). The metajob now has a little green dot in front of it, which means it is running:
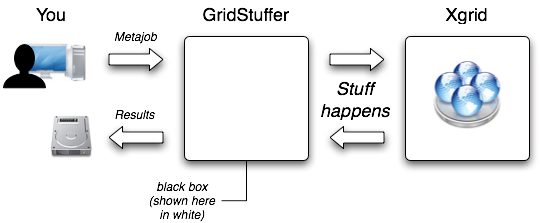
Stuff happens
You are actually done, and can go for a coffee, while GridStuffer runs. While drinking your coffee, have a look at how GridStuffer works at a higher level, both in terms of design and philosophy:
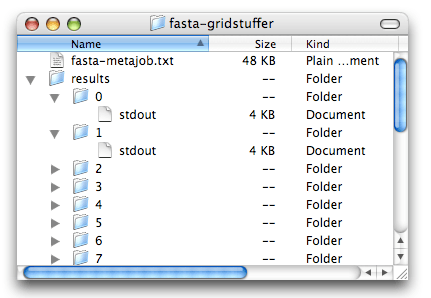
When the metajob is completed, all the results are available in the Results folder that you created earlier. The contents of this folder is shown in the screenshot below. For each command that we typed in our input file, GridStuffer created a folder: folder “0” for the first line, folder “1” for the second line,… We conveniently used the first line for chromosome X, which ends up in folder “0”, and the second line for chromosome 1, which ends up in folder “1”,… For instance, the results for chromosome 6 will be in folder “6”. These results consist of just one file, the output from the fasta command. Instead of displaying it on screen, GridStuffer saves it inside a file named “stdout” (short name for “standard output”). You can open these files in TextEdit to see the results, as shown below.

Validate the stuff
This tutorial won’t go into a detailed description of the most advanced features of GridStuffer (I will leave that for a future installment…). However, there is one feature that is worth mentioning, and that could make your life much easier, and this is “result validation”. To access this feature, select a metajob, and click on the ‘i’ icon in the MetaJobs window toolobar, to bring up the inspector for metajobs. There are a bunch of settings there, but the ones we will cover now are found under the “Validation” tab:
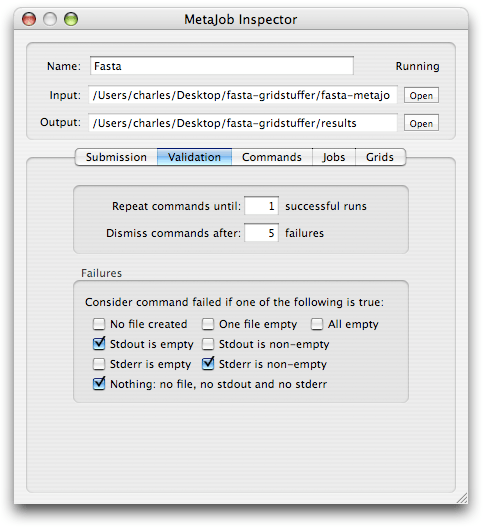
The first 2 items offer you control over the number of times a command will run on your grid:
- You could choose to run every command more than once, for instance twice to “double-check” the results, that might differ depending on the processor that the program will run on (G3? G5? Core Duo?). Maybe you even want to run each command several times, because there is some random component to the calculations, and you can apply some statistics to the different results.
- You could also choose to limit the number of “failures” a command can accept. When running a lot of computations, there is a greater chance that one of them will fail on a particular time and on a particular computer, for reasons that you did not anticipate or can’t control. However, when a particular command keeps failing and failing and failing again, you may want to give up after some time, like, after 5 failures. You can also leave this setting to 0, and the command will be submitted forever even if it keeps failing, and until all computers on earth have disappeared.
Importantly, GridStuffer will keep all results, and will never overwrite any file. When a command is run several times, you get all the results from the different runs. GridStuffer will also upload the results from failed tasks in a separate folder, so you can also check what went wrong.
But wait, there is more to be said on “failures”. In many cases, Xgrid will not necessarily know that things went wrong, because the problems happened at a level that Xgrid does not and should not have to care about. For instance, maybe you are running your job on an agent that has a corrupted fasta executable, and the job returns empty. Xgrid won’t necessarily notice that something went wrong and will consider the run to be successful. You may then have to look through thousands of results, and hand-pick the commands that failed, list them and finally run them again. And check the results again.
GridStuffer can help with that. For instance, with Fasta, you know that the standard output should not be empty (see results above) and when it is empty, you know something went wrong. You can then tell GridStuffer to consider a command “failed” if no ‘stdout’ is detected, by activating the corresponding check box (see screenshot above).
In general, GridStuffer will be able to look at these 3 different components in your results:
- The “standard output” or “stdout”. This is what a program would usually print on screen, such as the output from fasta.
- The “standard error” or “stderr”. This is usually empty, unless the program encounters an error. In the example of fasta, we don’t expect to have anything in the stderr stream, so we check the corresponding setting (see screenshot above).
- Files. Many programs also create additional files (tough fasta does not in this tutorial). For instance, your program may manipulate image files and save the modifications to disk. If no such file is created, or some of these files are completely empty, it is a sign that something went wrong, and GridStuffer should flag the corresponding run and consider it a failure.
End of the stuff
This is all for today’s tutorial… Next time, I will try to cover a more technical aspect of Xgrid, with multi-task job submissions, and how they are utilized in GridStuffer.
Leave a Reply